
| The degradation of plant biomass by the Trichoderma reesei enzyme cocktail: |
The development of alternative, low-carbon energies is essential in order to reduce greenhouse gas emissions and tackle climate change. One example is second-generation bioethanol, produced from lignocellullosic biomass and used as a fuel in the mobility sector.
My research, conducted over a period of more than 15 years at IFPEN and summarized in my HDR (accreditation to direct research), has focused on a crucial step in the production process for this biofuel: enzymatic hydrolysis1. Biomass degradation is carried out by a complex cocktail of enzymes produced by the filamentous fungus Trichoderma reesei. However, for industrial purposes, it is necessary to enhance the performance of this cocktail. It was to this end that I studied various aspects of its production, composition and improvement:
- Conditions for cellulase2 gene induction;
- Regulation of the secretion of these enzymes and identification of target genes to increase the fungus’ secretion capacity.
- Enzyme interaction with the lignocellulosic substrate at various stages of hydrolysis;
- Using a mathematical model, identification of limiting enzymes for a given substrate (limitation being due to their inadequate proportion in the cocktail and/or their insufficient activity). For the hydrolysis of wheat straw, the composition of a cocktail leading to maximized yield could be predicted;
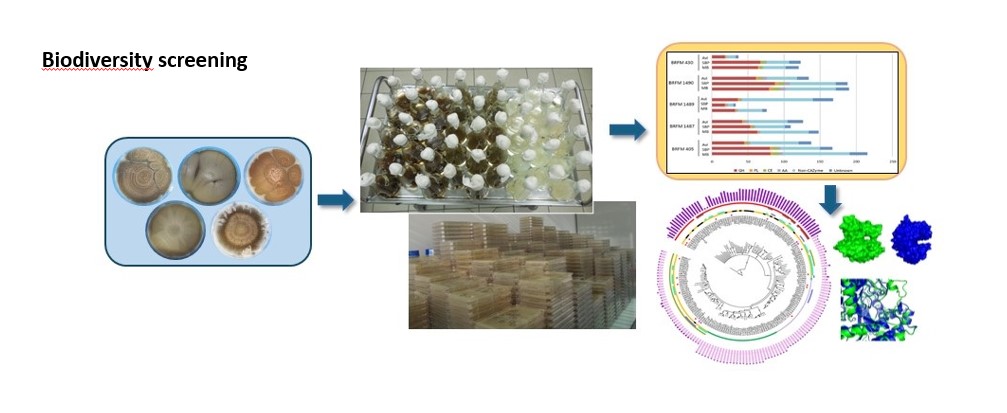
- Improvement of the cocktail via the addition of enzymes obtained from microbial biodiversity (see figure).
With respect to the latter point, the enzymes secreted (known as the “secretome”) by different filamentous fungi, such as Podospora anserina and Aspergillus japonicus, were analyzed. From the secretomes providing the biggest boost of activity when incorporated in the T. reesei cocktail, the enzymes potentially responsible for this improvement were identified using mass spectrometry. In one of these secretomes, AA16s, a new family of enzymes capable of boosting cellulases was discovered and characterized [1].
I also explored bacterial biodiversity within a metagenomic approach3. Initially, a functional screening of a gene library resulted in the identification of about forty enzymes acting on lignocellulosic biomass. Four enzymes belonging to a family of endoglucanases were then characterized and their mechanism of action on cellulose was described [2].
1 Degradation of a compound in the presence of enzymes following its reaction with water
2 Enzyme that hydrolyses cellulose
3 A method for studying the genetic content of samples taken from complex environments
This research opened up new avenues for progress, through the identification of limiting enzymes and targets for improving the secretion system in T. reesei. As a result, I demonstrated the potential of biodiversity exploration for the discovery of new efficient enzymes, and it is in this vein that I am currently pursuing my research.
References :
- Filiatrault-Chastel C, Navarro D, Haon M, Grisel S, Herpoël-Gimbert I, Chevret D, Fanuel M, Henrissat B, Heiss-Blanquet S, Margeot A, Berrin JG. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes.Biotechnol Biofuels. 2019; 12:55
>> https://doi.org/10.1186/s13068-019-1394-y
- Aymé L, Hébert A, Henrissat B, Lombard V, Franche N, Perret S, Jourdier E, Heiss-Blanquet S. Characterization of three bacterial glycoside hydrolase family 9 endoglucanases with different modular architectures isolated from a compost metagenome. Biochim Biophys Acta Gen Subj. 2021; 1865(5):129848
>> https://doi.org/10.1016/j.bbagen.2021.129848
To contact : Senta Blanquet






